The Details panel, represented by a blue “information” icon ( 

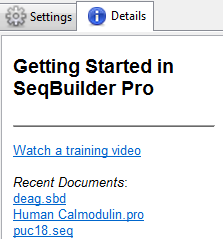
When nothing is selected, the Details panel displays links to help you get started in SeqBuilder Pro. Click on the links to watch a training video or open a recent document.
Otherwise, the Details panel will include a subset of the following information, depending on what is currently selected (e.g., a feature, sequence range, range between two enzymes).
| Statistic | Description |
|---|---|
| Selection and length | Whether the selection is on the forward or reverse strand and the endpoints of the selection. This is followed by the length of the selected region in base pairs. 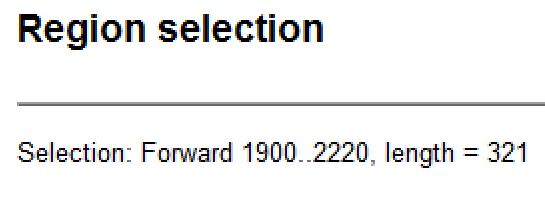 |
| Statistics | A table of statistics that may include the melting temperature (Tm) and the base composition of the selection. Among other dinucleotides, the analysis table may include the %CG dinucleotide. This dinucleotide is relevant to mammalian biology, as it is methylated in cells and clusters of CG are associated with certain types of promoters. |
| Enzyme information | When one or more enzymes are individually selected, the panel displays the name of each selected enzyme, along with a portion of sequence showing its cut sites on the top and bottom strands. If you select the range between two enzymes, this section shows the enzyme names and their cut sites.  |
| Feature Information | This section is compiled from annotations included in the sequence when it was opened (e.g., a Genbank file); or added later using the Features view. 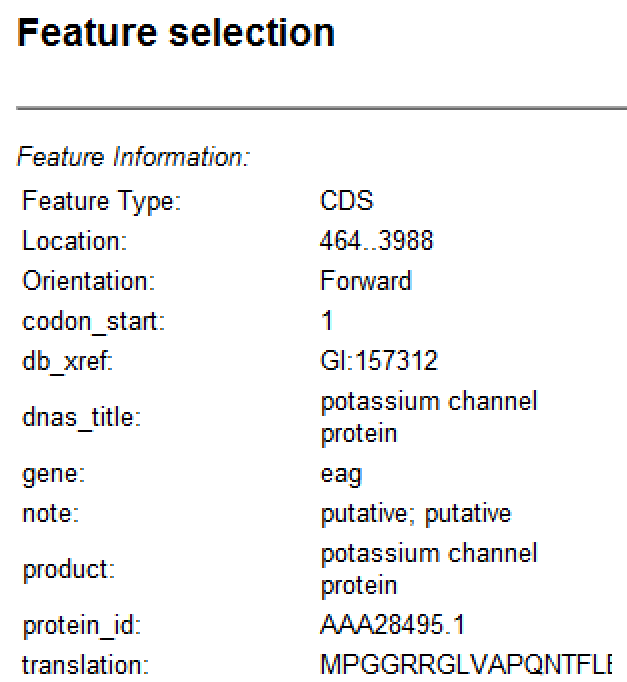 If multiple features are selected, the panel contains information about how many features are selected and how many are currently visible in the view.  The Selection table contains information calculated by SeqBuilder Pro:
|
Need more help with this?
Contact DNASTAR


