The Primers view is one of the ten views available in the Document window, and shares a sub-set of button tools with the other views. To make it the active view (or one of two active views), click on the Primers tab ( 
The Primers view is available for nucleotide sequence only, and displays tabular results from primer searches, including the primer sequence and its length, product (for primer pairs), melting temperature, and free energy (ΔG). For information on creating, editing or deleting primers and primer pairs, see Work with Primers.
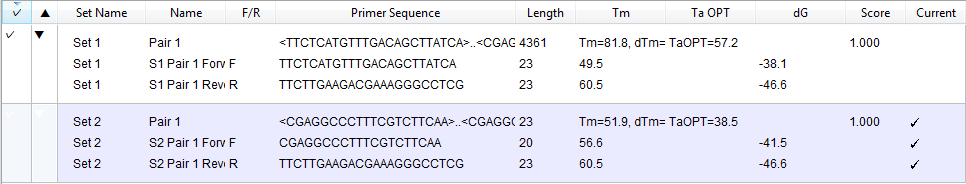
A checkmark next to a primer pair indicates that it is the currently selected pair. Select a row to also select that primer pair in the graphical views (Circular, Linear, Sequence). The table supports selection of groups of primer pairs using Ctrl/Cmd+click, Shift+click, or by dragging the mouse across the desired rows.
You can rename a primer by expanding its entry in this view (i.e. by clicking on the plus sign) and then clicking on a Name cell. Type in any desired name and press Enter.
The table below describes each of the columns in the Primers view.
| Column | Description |
|---|---|
| Show | A checkmark in this field denotes a feature that is currently visible in the Primer Design view, as well as the other graphical views (Circular, Linear, Sequence). To hide a feature, click its checkmark. |
| Active | A bullet point in this field indicates that the pair is the currently selected pair in the current working set. The current pair is always displayed in the Primer Design view. |
| Set Name | The name of the set. One set is created for each primer search. By default, SeqBuilder Pro assigns a generic name, like Set 1, Set 2, etc. However, this field is editable so that you can assign a more descriptive name for the set of primers, if desired. |
| Pair Name | The name of the primer pair. By default, SeqBuilder Pro assigns a generic name, like Pair 1, Pair 2, etc. However, this field is editable so that you can assign a more descriptive name for your primer pair, if desired. |
| F/R | An F in this column indicates that the primer is found on the forward (top) strand of the template sequence; an R indicates that it is on the reverse (bottom) strand. |
| Primer Sequence | For primer pairs, the sequences are shown for both members of the pair, forward strand followed by reverse strand. For primers, the sequence of the primer is shown in the 5’ to 3’ orientation, and is editable. Changes made to the primer sequence are automatically reflected in the Primer Design view. |
| Length | For primer pairs, this value indicates the length (in bp) of the PCR product. For primers, it indicates the length (in bp) of the primer. |
| Tm | For primers, the melting temperature of the primer is displayed. For primer pairs, 3 different values are listed: * L: The length of the product. * Tm: The melting temperature of the product. * dTm: The melting temperature difference between the product and the lower primer Tm. * TaOPT: The optimal annealing temperature for the primer pair. * dG: The delta-G value, free energy, that is computed over of the whole primer. Note that the dG value is only calculated for individual primers, not primer pairs. |
Need more help with this?
Contact DNASTAR


